New publication on the genomic diversity of Aspergillus fumigatus
Work from Amelia’s postdoc was recently published in Nature Microbiology.
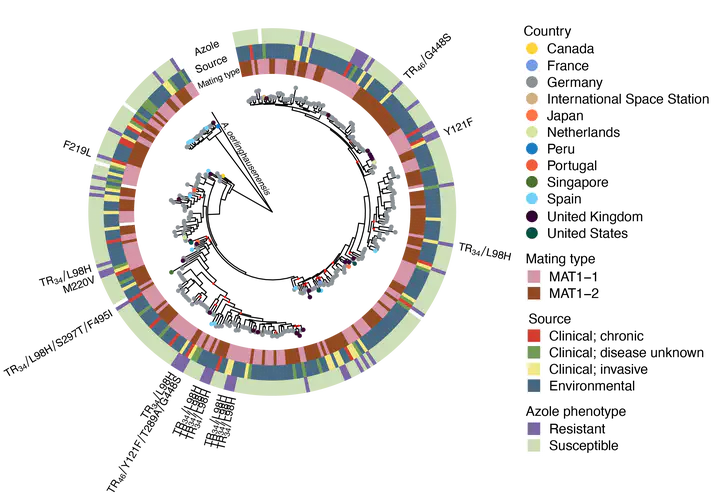
Aspergillus fumigatus is a fungus that is widespread in the environment and causes life-threatening infections in humans. Despite the more than 300,000 cases of invasive disease globally each year, a comprehensive survey of the genomic diversity present, including the relationship between clinical and environmental isolates, and how this genetic diversity contributes to virulence and antifungal drug-resistance, has been lacking. In this study, we define the pangenome of A. fumigatus using a collection of 300 genomes (83 clinical isolates and 217 environmental isolates) from a global distribution. We find that of the 10,907 unique orthogroups, 69% are core and found in all isolates, while 3,344 show presence/absence variation, representing 16-22% of each isolate’s genome. Using this large genomic dataset of environmental and clinical samples, we found an enrichment for clinical isolates in a genetic cluster whose genomes also contain more accessory genes, including more transmembrane transporters, proteins with iron-binding activity, and genes involved in both carbohydrate and amino acid metabolism. This characterization of the genomic diversity allows us to move away from a single reference genome that does not necessarily represent the species as a whole and better understand its pathogenic versatility, ultimately leading to better management of these infections.
The article is available as a PDF here or you can view the article at Nature Microbiology.
The press release can be found here in both English or German.